OpenMetadata is one of the up-and-coming open-source data catalogs which saw its first major release in April 2023. OpenMetadata was initially created by the engineers who worked on and learned from Uber’s Databook project to solve their data discovery and cataloging problems. OpenMetadata’s capabilities include integrating various tools from the modern data stack, such as messaging platforms, databases, and workflow engines.
Over the last few years, dbt has become the de facto standard for automating transformation workloads in data warehouses, data lakes, and lakehouses. dbt’s capability to transform data from one layer to another is backed by a metadata-driven approach. With many metadata, query templates, and dependencies, among other things defined in dbt, it has become quite an enticing data source for data catalogs. It can contribute to data asset enrichment and help with metadata lineage, orchestration, etc.
This article will take you through how OpenMetadata and dbt come together to provide you with an integrated view of your data assets, enriched and in one place.
OpenMetadata Ingestion and Workflows
Permalink to “OpenMetadata Ingestion and Workflows”OpenMetadata has a metadata ingestion framework that sets the foundation for all external connectors. The ingestion framework allows you to ingest:
- Core structural metadata using the Metadata Ingestion Workflow
- Data source usage metadata with Usage Workflow
- Data profiling metadata using the Profiler Workflow
- Data quality metadata using the Data Quality Workflow
- Data lineage metadata using the Lineage Workflow
- dbt metadata using the dbt Workflow
dbt was a part of the Metadata Ingestion Workflow until the 0.13.1 release. Now, dbt has its own separate workflow called the dbt Workflow. OpenMetadata’s Python-based workflow-agnostic ingestion framework allows for quick and easy development of new types of metadata to be ingested.
The ingestion framework uses MySQL as the durable storage, Elasticsearch as the search engine, and the OpenMetadata server for communicating with both these and other components of OpenMetadata. It is a flexible component that can be deployed from a Docker image independently or used as a Python connector, say, in Apache Airflow, Google Cloud Composer, or AWS MWAA.
With that in mind, let’s zoom in on how dbt Workflows help ingest different types of metadata that dbt offers.
OpenMetadata dbt Workflow overview
Permalink to “OpenMetadata dbt Workflow overview”OpenMetadata supports the ingestion of the metadata related to the following dbt features from dbt v1.2 onwards:
- Models - the transformation queries used in dbt Models.
- Lineage - the dependencies defined using the ref function and represented using a DAG.
- Tags - the general-purpose construct of metadata classification.
- Ownership - the information defined using the model owner construct.
- Descriptions - for models, tests, seeds, snapshots, analyses, macros, etc.
- Tests - the tests defined on models, seeds, snapshots, etc.
To get all of this data into OpenMetadata, you can use one of two methods:
- Use the OpenMetadata dbt ingestion UI, where it will ask you to populate the credentials of the storage account where your configuration files are stored-for instance, Amazon S3, Google Cloud Storage, on-prem file servers, dbt Cloud, etc.
- Create a configuration file with dbt as a source with the credentials of the files mentioned above and run the
metadata ingestcommand using the CLI.
Let’s look at some specifics of enabling the dbt Workflow ingestion in OpenMetadata.
OpenMetadata dbt Model ingestion
Permalink to “OpenMetadata dbt Model ingestion”Everything in dbt is represented using dbt Artifacts, of which the manifest and the catalog are of prime interest for the OpenMetadata use case. dbt Artifacts are produced when dbt is invoked to perform certain functions. For instance, the catalog.json file is produced when you run the docs generate command in the CLI.
The catalog.json file contains the data dictionary of all your dbt sources. The manifest.json contains your models, seeds, snapshots, tests, macros, and metrics, among many other things. You can use the run_results.json file for getting data for an invocation of dbt, i.e., running your dbt transformations.
Moreover, as you’ll see in the next section, OpenMetadata uses other mechanisms like the lineage parser to infer and ingest metadata from dbt.
OpenMetadata dbt Lineage ingestion
Permalink to “OpenMetadata dbt Lineage ingestion”The dbt Lineage ingestion works at two levels. Firstly, OpenMetadata reads the depends_on section for every node in your manifest.json file, an example of which is shown below:
{
"model.dwh.fact_sales": {
"compiled": true,
"resource_type": "model",
"depends_on": {
"macros": [],
"nodes": [
"model.dwh.dim_orders",
"model.dwh.dim_stores",
"model.dwh.dim_customers"
]
}
}
}
Another method OpenMetadata uses for fetching lineage metadata is to use the Lineage parser if the compiled variable is set to true in the node, also specified in the example above. The graphical representation of the lineage above would look something like the following:
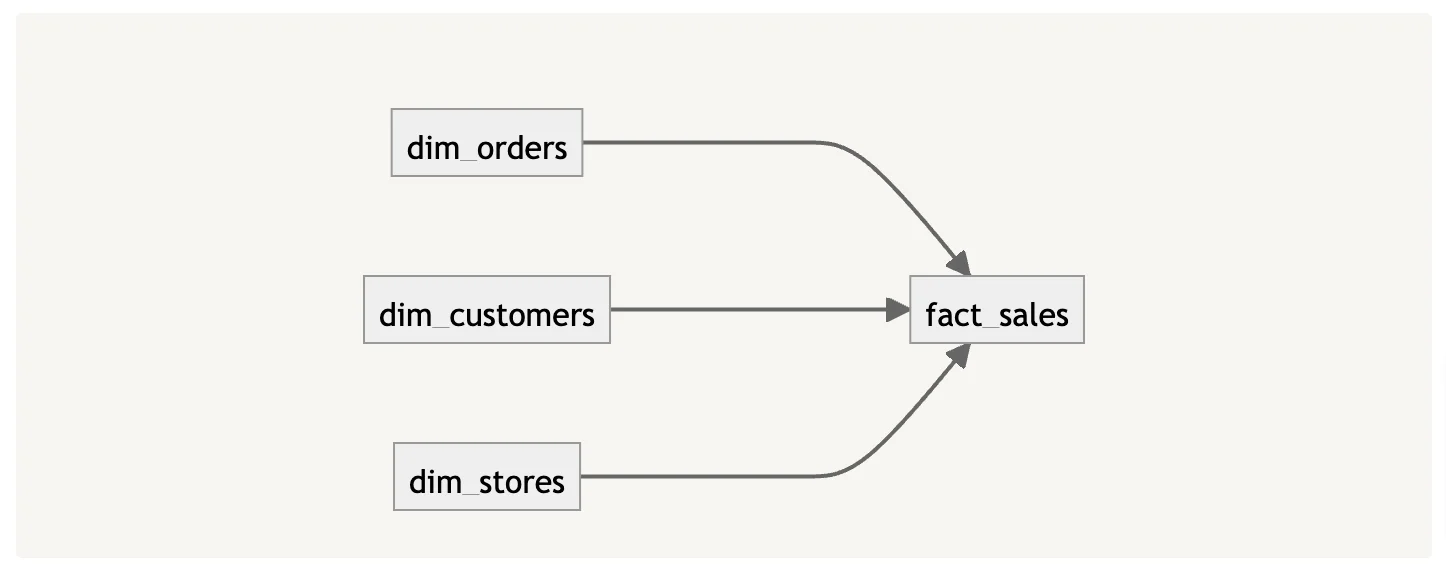
OpenMetadata dbt Lineage ingestion - Image by Atlan.
Using the lineage metadata, OpenMetadata reproduces the lineage graph within the UI, reducing the need to go to the dbt console to look at the lineage graph.
OpenMetadata dbt Tests ingestion
Permalink to “OpenMetadata dbt Tests ingestion”Whether you use dbt-native tests or packages like dbt_expectations, you can bring the execution results from those tests into OpenMetadata using the dbt Workflow. Test results are captured in the run_results.json file. OpenMetadata uses this file to ingest the test metadata.
Here’s a snippet from a sample result item from the run_results.json file:
{
"results": [
{
"status": "success",
"timing": [
{
"name": "compile",
"started_at": "2023–07–31T20:12:20.283068Z",
"completed_at": "2023–07–31T20:12:20.294936Z"
},
{
"name": "execute",
"started_at": "2023–07–31T20:12:20.143407Z",
"completed_at": "2023–07–31T20:12:20.145132Z"
}
],
"thread_id": "Thread-1",
"execution_time": 0.015143663541245722,
"adapter_response": {},
"message": null,
"failures": null,
"unique_id": "model.dbt_expectations_integration_tests.not_null_test"
}
]
}
Once the test results are ingested, they are visible under the Profiler & Data Quality tab on the OpenMetadataUI.
OpenMetadata dbt Tags ingestion
Permalink to “OpenMetadata dbt Tags ingestion”Tags are general-purpose labeling constructs to help you enable different workflows, roles, and functions, among other things. In the following example, two tags are defined on a table called dim_orders to signify that it is part of the core data model and also that it is a dimension type 2 table:
{
"model.dwh.dim_orders": {
"compiled": true,
"resource_type": "model",
"depends_on": {},
"tags": [
"dim_type_2",
"core_model"
]
}
}
Once the metadata from dbt flows into OpenMetadata, efficient tagging helps tremendously with search and discovery. Multi-tag searches can narrow down the search results to a high degree. Different teams can have tags specific to their use cases and workflows, reducing the search area further. Sometimes tags aren’t enough; you need to look at a more detailed description of your data assets, which is where data asset descriptions come in. Let’s talk about them in the next section.
OpenMetadata dbt Descriptions ingestion
Permalink to “OpenMetadata dbt Descriptions ingestion”OpenMetadata allows you to ingest the descriptions of tables and columns defined in the manifest.json and catalog.json files in dbt. Only descriptions provided in the manifest.json file are ingested by default. Here’s an example of descriptions of a table and column provided in the model’s YAML file:
version: 2
models:
- name: dim_orders
description: "Dimension table containing orders from all business units"
columns:
- name: order_value
description: "Actual realized value of the order"
Once the descriptions are ingested, OpenMetadata follows the usual flow where the ingestion framework calls the OpenMetadata backend, which, in turn, writes the data to the MySQL backend, the Elasticsearch search engine, and feeds the UI directly.
OpenMetadata dbt Ownership ingestion
Permalink to “OpenMetadata dbt Ownership ingestion”You can define an owner for all your dbt assets. The ownership information gets stored in the manifest.json and the catalog.json files. To get the ownership information into OpenMetadata, you must ensure that the owner defined in dbt is also a user (or a team) in OpenMetadata.
If you don’t already have that user, you’ll need to manually create the user that corresponds to the owner’s email using the OpenMetadata UI. As mentioned above, objects can also be owned by teams. In those cases, you’ll need to manually create a team using the OpenMetadata UI. Here’s how your owner will be defined in dbt for the dim_orders table model:
{
"model.dwh.dim_orders": {
"metadata": {
"type": "dimension type 2 table",
"schema": "dwh",
"name": "dim_orders",
"database": "staging",
"owner": "data_engineering"
}
}
}
As dbt Workflow is a reasonably new addition to the ingestion workflows in OpenMetadata, you’ll be able to ingest more types of metadata with greater flexibility into OpenMetadata.
Summary
Permalink to “Summary”This article showed how OpenMetadata and dbt work together to bring you the power of metadata, enabling many different dbt-powered features without leaving the OpenMetadata UI. This is of great use to the teams using OpenMetadata as their core data discovery and cataloging tool.
You can find more information about dbt on their official blog and about OpenMetadata on theirs.
Share this article



















